|
I am a third-year PhD student in Bioengineering at Caltech, where I study collective behaviors of active biomolecules. My PhD advisor is Matt Thomson. From 2019 to 2021, I worked in the Department of Data Science at Dana-Farber Cancer Institute and Systems Biology at Harvard Medical School with Sahand Hormoz. I primarily worked on developing single-cell and DNA technologies to investigate cell differentiation and cancer development. I did my undergraduate studies in Mechanical Engineering and Aerospace Engineering at CWRU, where I did research on sickle cell disease in Umut Gurkan's lab. sliu7[at]caltech.edu / Google Scholar / Twitter / GitHub |

|
|
My research interests are in the intersection of synthetic biology and physics. Both living and non-living active matters create non-equilibrium processes in which many individual parts consume energy and collectively generate motion and force. Active matters exhibit many interesting properties, and I am most interested in the phase transitions between local and global/collective behaviors. We use components from the cytoskeleton, mainly motor proteins and microtubules, as a model system to decode non-equilibrium dyanmics of active matters. I use theortical and experimental tools to investigate how the flow of time and the "greed" of nature evolve or alter active materials. I aim to: (1) understand and control the transition between local and global behaviors, (2) bridge our unsderstanding of biomolecules from cytoskeleton and mechanisms of neurodegenerative diseases, and (3) design programmable active material using a number of novel biomolecules. Aside from active matters, I'm also interested in developmental biology and biomechanics. I previously worked on developing single-cell technologies to measure cell states in diverse biological systems (mainly on bacterial and mammalian cells). I also worked on building organoid systems based on understanding obtained from our high resolution cell state data.For an up-to-date and full publication list, please see my Google Scholar page. (*equal contribution) |
|
|
|
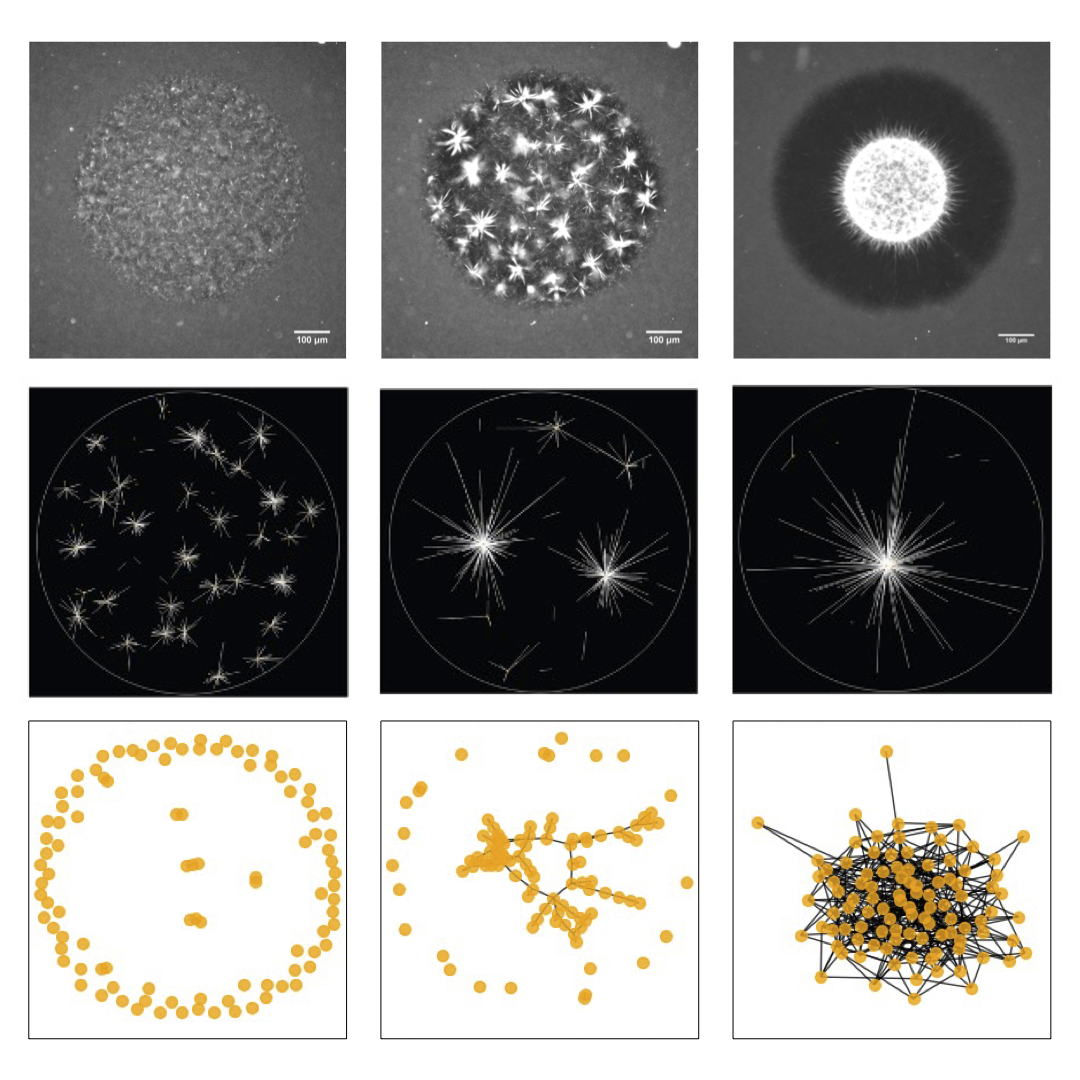
Active-molecule-induced movements are responsible for locomotion, reconfiguration, and replication of living systems. Experimental active-matter systems consist of motor proteins and microtubules are capable of multiscale self-organization ranging from contractile and extensile phases with millimeter-scale behaviors to local aster phases that only span a few micrometers. However, connecting the large-scale organization behavior of active materials to microscopic interactions between motors and microtubules remains a challenge due to lack of understanding on the multiscale structure of such systems. Here we uncover the micrometer- and millimeter-scale phase transition phenomena by mediating the bundling effect of microtubules. Combining experiments and simulations, we demonstrate that microtubule bundling facilitates transitions between local and global phases of organization. In our experimental system, we observe an increase in spatial correlation length between active molecules as the effective length of the microtubule structures increase. This provides evidence for how long-range organization can be generated when local molecular interactions reach a sufficient length scale. We anticipate our system to be a starting point for more sophisticated frameworks for both cellular organization and control engineering of multiscale collective behavior. |
|||||||||||||||
|
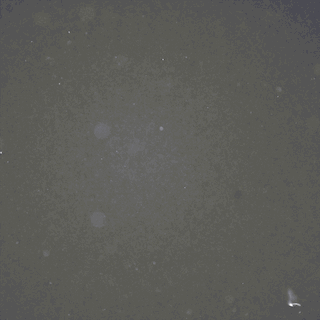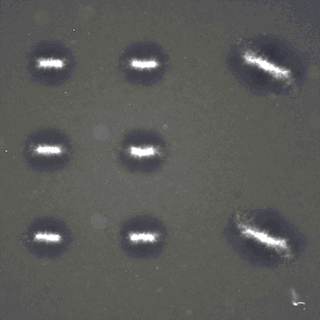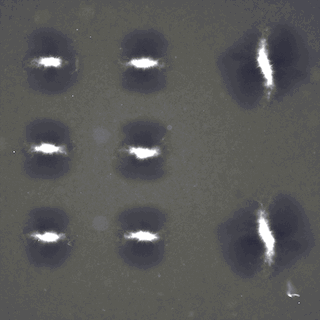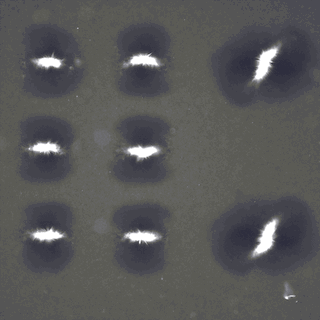
Active fluids with such living matter can be chaotic without a control mechanism, limiting their utility for bioengineering applications. Recently-developed light-activatable motors, which only crosslink the microtubules under illumination, have been previously applied to sculpt persistent fluid flows. Combining theoretical predictions and experimental results, we find that the linear superimposition can be achieved above a critical distance. Therefore, different flows can be realized simultaneously within a single channel through the spatio-temporal composition and manipulation of light patterns, making multitasking possible in a single channel. Our theoretical framework may be extended for other emergent structures of cytoskeleton, and our platform has potentials for building optically programmable flow-control devices. |
|
|
|
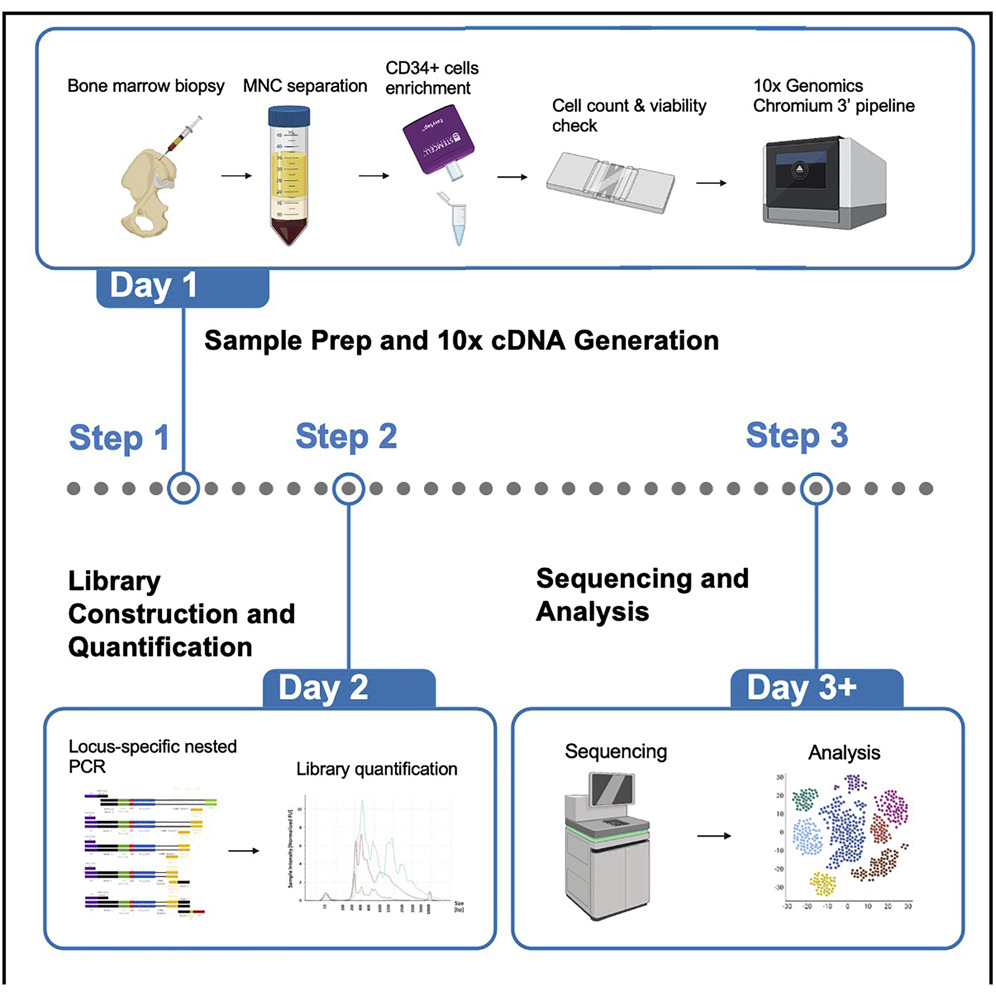
In many biological applications, the readout of somatic mutations in individual cells is essential. For example, it can be used to mark individual cancer cells or identify progenies of a stem cell. Here, we present a protocol to perform single-cell RNA-seq and single-cell amplicon-seq using 10X Chromium technology. Our protocol demonstrates how to (1) isolate CD34+ progenitor cells from human bone marrow aspirate, (2) prepare single-cell amplicon libraries, and (3) analyze the libraries to assign somatic mutations to individual cells. |
|||||||||||||||
|
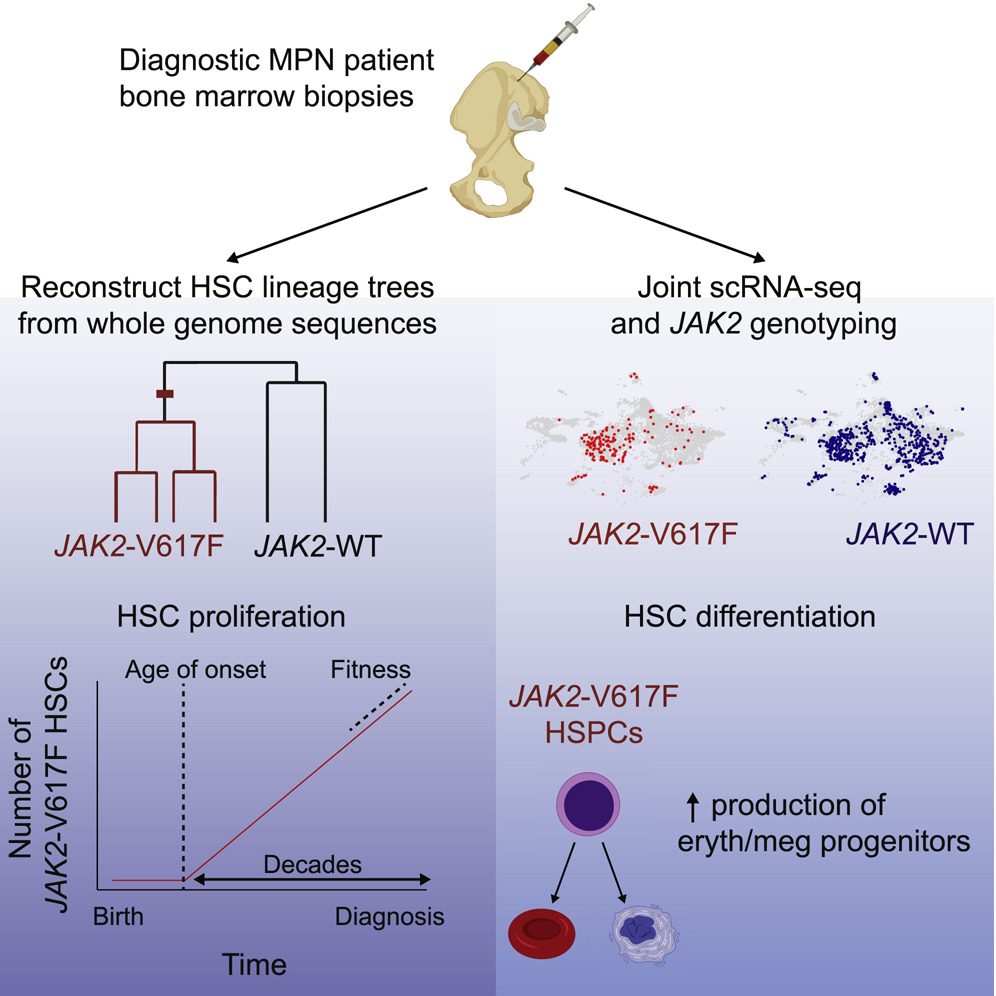
Blood cancers known as myeloproliferative neoplasms (MPNs) are thought to originate when a driver mutation is acquired by a hematopoietic stem cell (HSC). Here we quantified the effect of the JAK2-V617F mutation on the self-renewal and differentiation dynamics of HSCs in treatment-naive individuals with MPNs and reconstructed lineage histories of individual HSCs using somatic mutation patterns. We found that JAK2-V617F mutations occurred in a single HSC several decades before MPN diagnosis and found that mutant HSCs have a selective advantage. These results highlight the potential of harnessing somatic mutations to reconstruct cancer lineages. |
|||||||||||||||
|
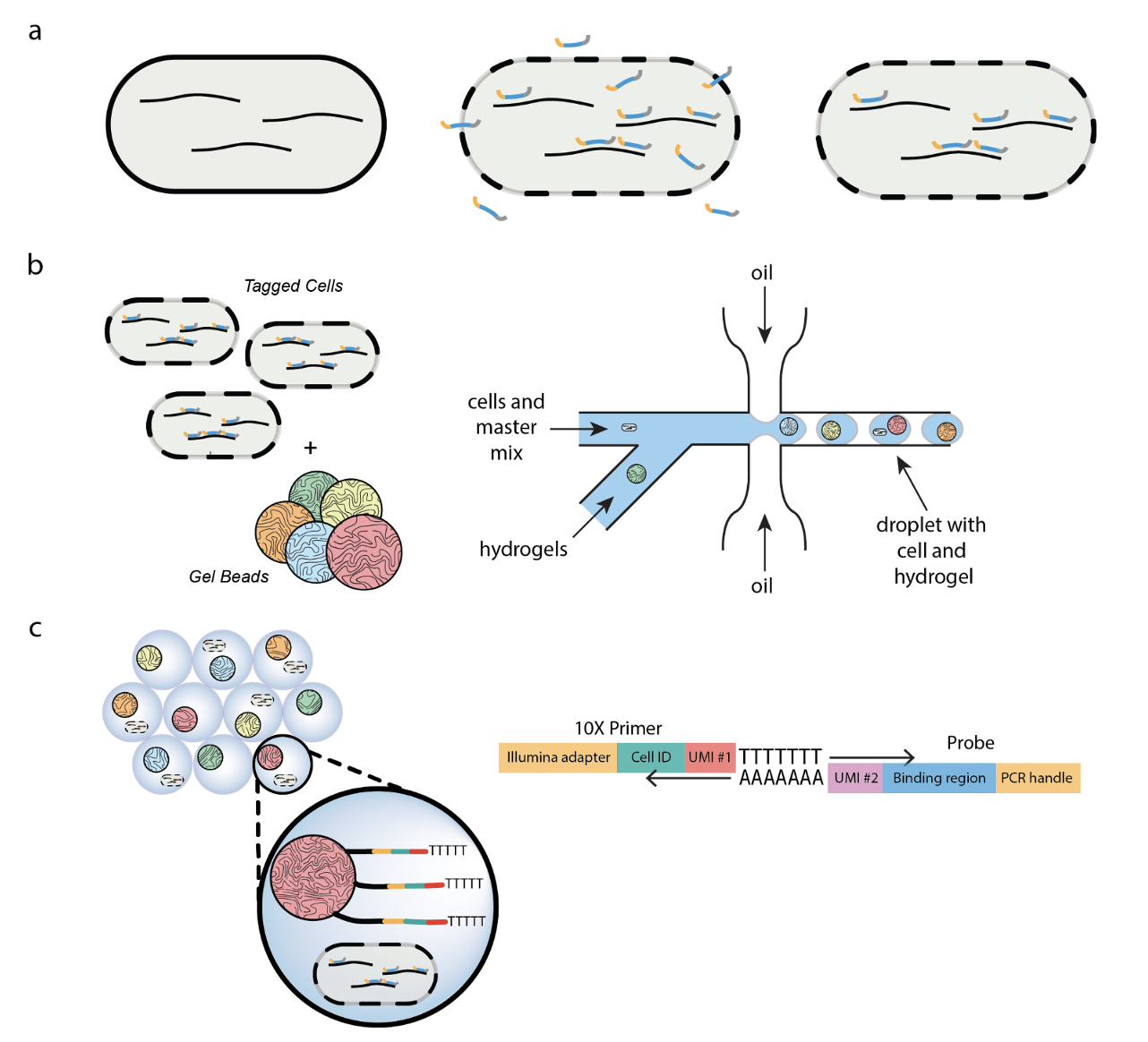
Here, we develop a method that uses DNA probes and leverages an existing commercial microfluidic platform (10X Chromium) to conduct bacterial single cell RNA sequencing. We sequenced the transcriptome of over 15,000 individual bacterial cells, detecting on average 365 transcripts mapping to 265 genes per cell in B. subtilis and 329 transcripts mapping to 149 genes per cell in E. coli. Our findings correctly identify known cell states and uncover previously unreported cell states. |
|
|







